7.2. Reading Osmose parameters
The loading of Osmose parameters is achieved by using the read_osmose function with an input argument providing the path of the
main configuration file.
suppressMessages(require("osmose"))
output.dir = file.path("rosmose", "_static")
exampleFolder <- tempdir()
demoPaths <- osmose_demo(path = exampleFolder, config = "eec_4.3.0")
# load the Osmose configuration file
osmConf = readOsmoseConfiguration(demoPaths$config_file)
# Summarize the configuration
summary(osmConf)
# extracting a particular configuration object
sim = get_var(osmConf, what="simulation")
# proper way to recover the season parameter (returns a vector of numeric)
movements = get_var(osmConf, what='movement')
# Plotting reproduction growth
png(filename = file.path(output.dir, 'species.png'))
plot(osmConf, what="species", species=0)
# Plotting reproduction seasonality
png(filename = file.path(output.dir, 'reproduction.png'))
plot(osmConf, what="reproduction", species=1)
# Plotting size range for predation
Length Class Mode
osmose 2 -none- list
mortality 4 -none- list
simulation 7 -none- list
fishing 1 -none- list
movement 7 -none- list
fisheries 9 -none- list
predation 4 -none- list
reproduction 1 -none- list
species 17 -none- list
grid 3 -none- list
population 1 -none- list
output 19 -none- list
Furthermore, some parameters can be plotted for a given species. This is done as follows:
png(filename = file.path(output.dir, 'predation.png'))
plot(osmConf, what="predation", species=2)
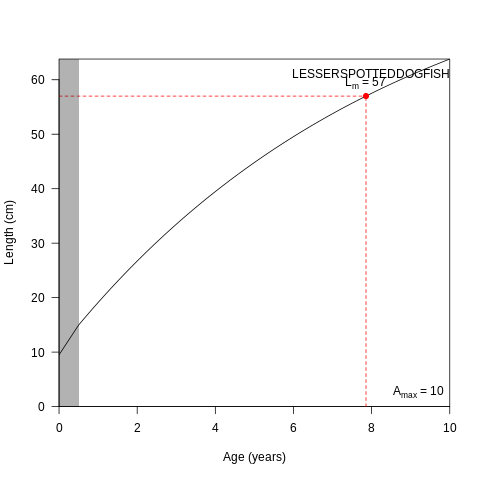
Fig. 7.1 Growth parameters for species 0
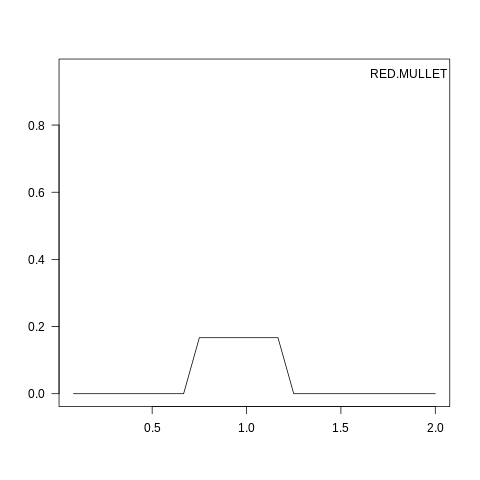
Fig. 7.2 Reproduction seasonality for species 1
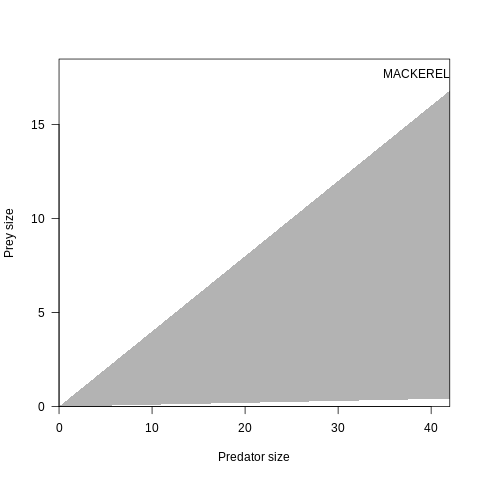
Fig. 7.3 Predation size range for species 2